|

German Version
Site map
FAIR 3889
Virus diseases
Phytoplasma diseases
Pathogen collection
Pathogen detection
ACLSV
ApMV
ASGV
ASPV
PPV
PDV
PNRSV
ArMV
ToRSV
RpRSV
SLRSV
GFLV
GLRaV-1
GLRaV-3
LChV
CMLV
CRMV
CNRMV
CGRMV
ChTLV
CVA
AP
ESFY
PD
Pathogen elimination
Contact us
Related Sites
|
Pathogen detection
Different methods for the detection of plant pathogens have been developed
and are applied, depending on the pathogen to be detected, the time frame,
equipment and financial ressources available. These aspects are important
for the choice of a test system, when to be applied for traded budwood.
Internationally approved and recognized detection systems including serological
and molecular laboratory assays and indicator hosts in greenhouse and field indexing
were updated regularly by the ISHS International Working Group on Fruit Tree
Viruses in occasion of their meetings 1991 in Vienna (Acta Hort. 309: 407-418,
1992), 1997 in Bethesda (Acta Hort. 472: 761-783, 1998) and 2000 in
Canterbury (Acta Hort. in press).
An overview of these methods for pome fruit and
stone fruit pathogens is given in the tables at the bottom
of this page.
Viruses
Viruses can be detected by field and greenhouse
indexing,
serology, optical methods like electron mircoscopy,
molecular hybridization, Nucleic Acid Sequence Based Amplification (NASBA)
and PCR amplification.
The ISHS ´Working Group on Virus Diseases of Fruit Trees´
recommends serological indexing (ELISA) since many years. Since the Bethesda meeting
in 1997 major emphasis is layed on the application of PCR based detection
systems, due to their increased sensitivity and velocity.
In the frame of FAIR 3889 improved protocols both for RNA preparation but
also for specific and broad range detection of fruit tree viruses have been
established.
Back to pagetop
Phytoplasmas
Phytoplasmas may be detected by orchard and nursery inspections due to
typical morphological anomalies. ESFY can be reliably detected by indexing
on GF 305 seedlings in the greenhouse with an incubation period of
approximately 4 months. In the sieve tubes of petioles, bark and roots
phytoplasmas can be detected by
DAPI staining.
In the frame of FAIR 3889 PCR was optimized as rapid and
reliable diagnostic tool. The PCR detection of phytoplasmas relies on the
quality of the DNA template for amplification. Therefore different protocols
for DNA preparation are recommended. Plate Capture PCR or IC-PCR avoids an
initial preparation step by use of antisera.
Back to pagetop
Indexing
Indexing is based on the ability of special indicator plants to develop
typical disease symptoms after infection with a certain virus. The cultivar
to be tested is grafted to the indicator or plant sap is trasmitted mechanically.
After a given incubation time the presence of visible symptoms is determined.
The following tables give an overview of approved indexing methods and the
viruses that may be detected on indicator plants.
Further information about indexing is available on the homepage of
the Washington
State University.
| Type | Indicator | Duration |
Detectable Viruses |
|---|
| Herbaceous Greenhouse Indexing |
Chenopodium quinoa | 20 days |
ACLSV, ASGV, Nepoviruses |
| Cucumis sativus | 20 days |
ApMV, PDV, PNRSV |
| Woody Greenhouse Indexing |
Malus platycarpa | 8 weeks |
ACLSV |
| Malus pumila ´Virginia Crab´ | 24 weeks |
ASGV, ASPV |
| Malus pumila R 12740 7A | 4 weeks |
ACLSV |
| Malus pumila spy 227 | 12 weeks |
ASPV |
| Cydonia oblonga C 7/1 | 5 weeks |
ACLSV |
| Prunus persica GF 305 | 8 weeks |
ACLSV, PPV, PDV, PNRSV, SLRSV, ESFY |
| Prunus tomentosa | 12 weeks |
ACLSV, PPV, PDV, PNRSV |
| Prunus serrulata ´Shirofugen´ | 8 weeks |
PDV, PNRSV |
| Field indexing |
Malus platycarpa | 2 years |
ACLSV |
| Pyronia veitchii | 2 years |
ASPV |
| Malus pumila ´Virginia crab´ | 3 years |
ASGV, ASPV |
| Malus pumila R 12740 7A | 2 years |
ACLSV |
| Malus pumila spy 227 | 2 years |
ASPV |
| Malus pumila ´Lord Lambourne´ | 3 years |
ApMV, rubbery wood, flat limb, chat fruit |
| Malus pumila ´Gravensteiner´ | 3 years |
flat limb |
| Malus pumila ´Golden Delicious´ | 2 years |
ApMV, AP |
| Prunus serrulata ´Shirofugen´ | 6 weeks -
2 years |
PDV, PNRSV |
| Prunus serrulata ´Kwanzan´ | 2 years |
CGRMV |
| Prunus avium ´Bing´ | 3 years |
CRLV, CTLV, SLRSV, ArMV, CNRSM, CRMV |
| Prunus avium ´Sam´ | 3 years |
LChV |
| Prunus avium Canindex I | 3 years |
CNRMV |
| Prunus persica GF 305 | 4 years |
ACLSV, PPV, PDV, PNRSV, ESFY |
Back to pagetop
Serology
The ELISA (Enzyme Linked Immunosorbant Assay) test has been developed for the
detection of plant viruses almost
30 years ago. It is used routinely for large scale testing of plants for the
presence of many viruses. It is rapid, inexpensive and convenient. However,
ELISA can only be applied for those viruses where specific antisera are
commercially available.
It is further limited by the eneven patterns of distribution of certain pathogens
in a tree, but also by climatic influence, reducing the titer below the
possible level of detection.
In sanitation programmes after elimination treatments it may therefore require
some time before virus replication reaches again a detectable treshold.
The use of Immuno-Tissue-Printing allows the localization of viruses in tissues
and therefore the improvement of elimination strategies in vitro.
Back to pagetop
Polymerase Chain Reaction (PCR)
Polymerase Chain Reaction is a method for amplification of specific
DNA regions, producing easily detectable amounts of DNA fragments, which
are usually visualized by agarose gel electrophoresis.
Concerning pathogen detection, PCR is 100- to 1000-fold more sensitive than ELISA.
Therefore it is specially suitable for the detection of infections in an initial
stage, where the pathogen titer in the plant is still low.
Many descriptions of the PCR technique exist in the World Wide Web. Following you find
a few links about this topic:
"What the heck is PCR" by C. Brown
"Principle of PCR" by Andy Vierstraete
Short description of PCR from the University of Califormia
Detailed PCR protocols from the University College in London
While the DNA of phytoplasmas can be used directly as a PCR template,
most plant viruses contain RNA. For their detection either an RNA
purification or, in case of pathogens for which antisera are available,
an immunocapture (IC) followed by an RT step is required to convert the viral
RNA to a DNA that can be amplified by PCR.
For pathogen detection PCR is carried out by using a range of
general or specific primer pairs which represent short DNA patterns of
the template DNA and flank the amplified region. Primers for
several important fruit tree pathogens where developed in the frame of FAIR 3889.
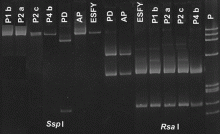
PCR products can be further analyzed in RFLP by enzymatic digestion. The resulting
´fingerprints´ confirm the identity of the amplified products and
may be used to identify sub-strains of a certain pathogen.
Back to pagetop
Serological and molecular laboratory assays available
for the detection of viruses and phytoplasmas in pome fruits
Pathogen
(click for detection details) |
Serological Assay | Molecular Assay | Reference |
|---|
| ACLSV | ELISA |
RT-PCR
IC-RT-PCR
RT-PCR ELOSA |
6, 27, 30, 31, 41 |
| ASGV | ELISA |
RT-PCR
IC-RT-PCR
RT-PCR ELOSA |
9, 27, 29, 30, 31, 37, 39 |
| ASPV | |
RT-PCR
NASBA |
25, 29, 30, 31, 38, 40, 58 |
| ApMV | ELISA |
RT-PCR
IC-RT-PCR
RT-PCR ELISA |
5, 56 |
| ToRSV | ELISA |
RT-PCR
RT-PCR ELISA
Hybridization |
18, 19, 43, 56 |
| AP | ELISA |
PCR
IC-PCR |
8, 21, 35, 36, 59 |
| PD | |
PCR |
8, 10, 12, 35, 36, 59 |
Back to pagetop
Serological and molecular laboratory assays available
for the detection of viruses and phytoplasmas in stone fruits
Pathogen
(click for detection details) |
Serological Assay | Molecular Assay | Reference |
|---|
| ACLSV | ELISA |
RT-PCR
IC-RT-PCR |
6, 27, 30, 31, 41 |
| ApMV | ELISA |
RT-PCR
IC-RT-PCR
RT-PCR ELISA |
5, 56 |
| PPV | ELISA |
RT-PCR
IC-RT-PCR |
|
| PDV | ELISA |
RT-PCR
IC-RT-PCR |
42 |
| PNRSV | ELISA |
RT-PCR
RT-PCR ELISA |
5, 20, 60 |
| ToRSV | ELISA |
RT-PCR
RT-PCR ELISA
Hybridization |
18, 19, 43, 56 |
| ArMV | ELISA |
RT-PCR
Hybridization
NASBA |
19, 37 |
| RpRSV | ELISA |
NASBA |
|
| SLRSV | ELISA |
NASBA |
|
| LChV | |
RT-PCR |
26, 28, 46, 49, 50, 53, 62 |
| CMLV | ELISA |
|
|
| CRLV | ELISA |
|
|
| CVA | |
RT-PCR
IC-RT-PCR |
22 |
| TRSV | ELISA |
|
|
| TBRV | ELISA |
|
|
| ESFY | |
PCR |
1, 8, 11, 13, 21, 23, 32, 35, 59 |
Back to pagetop
References
- Ahrens U., Lorenz K. H., and Seemüller E. 1993. Genetic diversity among
mycoplasmalike organisms associated with stone fruit diseases. Molecular
Plant-Microbe Interactions, 6: 686-691.
- Bertaccini A., Carraro L., Davies. D.L., Laimer da Câmara Machado M.,
Martini M., Paltrinieri S., and Seemüller E., 2000. Micropropagation of a collection
of phytoplasma strains in periwinkle and other host plants. 13th International
Congress of IOM, ACROS Fukuoka, Japan, July 14-19 2000: 101.
- Bertaccini A., L. Carraro, D. Davies; M. Laimer da Câmara Machado, M.
Martini, S. Paltrinieri, and Seemüller E., 2000. Micropropagation of a collection
of phytoplasma strains in periwinkle and other host plants. EFPP 2000,
Taormina-Giardini Naxos, 19-22 September 2000, 122P: 38.
- Candresse, T., 2001. Advances in the methods of pathogen detection.
Acta Hort. accepted.
- Candresse T., Kofalvi S. A., Lanneau M., and Dunez J., 1998. A PCR-ELISA
procedure for the simultaneous detection and identification of prunus necrotic
ringspot and apple mosaic ilarviruses. Acta Hort. 472.
- Candresse T., Lanneau M., Revers F., Grasseau N., Macquaire G., German S.,
Malinowsky T., and Dunez J., 1995. An immunocapture PCR assay adapted to the
detection and the analysis of the molecular variability of the apple chlorotic
leafspot virus. Acta Hort. 386: 136-147.
- Candresse, T., Lanneau, M., Revers, F., Kofalvi, S. and Macquaire, G., 2000.
PCR-based techniques for the detection of plant viruses and viroids.
Acta Hort. 530: 61-67.
- Carraro L., Nemchinov L., and Hadidi A., 1998. PCR detection of pome and
stone fruit phytoplasmas from active or dormant tissue. Acta Hort.
472: 731-735.
- Crossley S. J., Jacobi V., and Adams A. N., 1998. IC-PCR amplification of
apple stem grooving virus isolates and comparison of polymerase and coat protein
gene sequences. Acta Hort. 472.
- Davies D.L., Clark M.F. and Adams A.N. 1998. The epidemiology of Pear decline
in the UK. Acta Hort. 472: 669-672.
- Davies DL, and Adams AN., 2000. European stone fruit yellows phytoplasmas
associated with a decline disease of apricots in southern England. Plant Pathology
(in press).
- Davies DL., and Clark MF., 1994. Maintenance of MLOs occurring in Pyrus species
by micropropagation and their elimination by tetracycline therapy. Plant
Pathology 43: 819 - 823.
- Davies D. L., and Clark M. F., 1992. Production and characterisation of
polyclonal and monoclonal antibodies against peach yellow leafroll MLO-associated
antigens. Acta Hort. 309: 275-283.
- Foissac X., Svanella-Dumas L., Gentit P., Dulucq M.J., and Candresse T., 2001.
Polyvalent detection of fruit tree tricho, capillo and foveaviruses by nested
RT-PCR using degenerated and inosine containing primers (PDO-RT-PCR)
Acta Hort. accepted.
- Gentit P., Delbos R.P., Candresse T., and Dunez, J., 2001. Characterisation of
a new Nepovirus infecting apricot in the Southeastern France: Apricot latent
ringspot virus. Eur. J. Plant Pathol. accepted.
- Gentit P., Foissac X., Svanella-Dumas L., Peypelut M., and Candresse, T., 2001.
Biological properties and molecular characterization of two different Foveaviruses
inducing similar disorders in cherry trees. Acta Hort. accepted.
- Gentit P., Foissac X., Svanella-Dumas L., and Candresse T., 2001. Variants of
Apricot latent foveavirus (ALV) isolated from south european orchards associated
with peach asteroid spot and peach sooty ringspot. Acta Hort. accepted.
- Griesbach J. A., 1995. Detection of tomato ringspot virus by polymerase
chain reaction. Plant Dis. 79: 1054-1056.
- Hadidi A., and Hammond R. W., 1989. Construction of molecular clones for
identification and detection of tomato ringspot and arabis mosaic viruses. Acta
Hort. 235: 223-230.
- Hammond R., Howell W. E., Mink G. I., and Crosslin J. M., 1998.
Strain-specific polymerase chain reaction assays for discrimination of prunus
necrotic ringspot virus isolates. Acta Hort. 472.
- Heinrich M., Simona Botti, Licia Caprara, Arthofer W., Strommer S., Hanzer V.,
Paltrinieri S., Martini M., Katinger H. Bertaccini A., and Laimer da Câmara
Machado M., 2001. Development and evaluation of improved detection methods for
phytoplasmas in fruit tree. In preparation.
- James D., and Jelkmann W., 1998. Detection of cherry virus A in Canada
and Germany. Acta Hort. 472.
- Jarausch W., Lansac M., Saillard C., Broquaire J.M. and Dosba F., 1998. PCR
assays for specific detection of European stone fruit yellows phytoplasmas and its
use for epidemiological studies in France. European Journal of Plant Pathology
104: 17-27.
- Jelkmann W., 1998. Identification and detection of recalcitrant temperate
fruit crop viruses using dsRNAs and diffusion antisera. In: Plant Virus
Disease Control, A. Hadidi, R. K. Khetarpal, and H. Koganezawa, eds. APS
Press, St. Paul, MN, pp. 392-398.
- Jelkmann W., and Keim-Konrad R., 1997. An immuno-capture polymerase chain
reaction and plate-trapped ELISA for the detection of apple stem pitting virus.
J. Phytopathol. 145: 499-504.
- Jelkmann W., Keim-Konrad R., Vitushkina M., and Fechtner B., 1998.
Complete nucletotide sequences of little cherry closterovirus and virus
detection by polymerase chain reaction. Acta Hort. 472.
- Kinard G. R., Scott S. W., and Barnett O. W., 1996. Detection of apple
chlorotic leaf spot and apple stem grooving viruses using RT-PCR.
Plant Dis. 80: 616-621.
- Kountrias A., Eppler A., Rott M. E., Jelkmann W., and Adam, G. (2000).
Preliminary field results on two causal agents of little cherry disease in the
fruit growing area 'Altes Land' of Northern Germany. Acta Hort. (in press).
- Kummert J., Marinho V.L.A., Rufflard G., Colinet D., and Lepoivre P., 1998.
Sensitive detection of apple stem grooving and apple stem pitting viruses from
infected apple trees by RT-PCR. Acta Hort. 472: 97-104.
- Kummert J., Vendrame M., Steyer S., and Lepoivre P., 2000. Development of
routine RT-PCR tests for certification of fruit tree multiplication material.
EPPO Conference on Diagnostic Techniques for Plant Pests (Wageningen,
NL - 2000-02-1/4) Bulletin OEPP/EPPO Bulletin, 30.
- Kummert J., Vendrame M., Lepoivre P., and Steyer S., 2001. Development of
routine RT-PCR ELOSA tests for fruit tree certification. 18th International
Symposium on virus-like didease of temperate fruit crops - ISHS (Canterbury,
UK - 2000-07-9/15) Acta Hort. in press
- Laimer da Câmara Machado M., Paltrinieri S., Hanzer V., Arthofer W.,
Strommer S., Martini M., Pondrelli M. and Bertaccini A., 2001. Presence of
European stone fruit (ESFY or 16SrX-B) phytoplasmas in apricots in Austria.
Plant Pathology 50 (1) 130 - 135.
- Laimer da Câmara Machado M., Heinrich M., Hanzer V., Arthofer W.,
Strommer S., Paltrinieri S., Martini M., Bertaccini A., Kummert J. and Davies
D.L. 2001. Improved detection of viruses and phytoplasmas in fruit tree tissue
cultures. 18th ISHS Conference on Virus and Virus-like Diseases of Temperate
Fruit Crops. Canterbury, 9. - 15. July 2000. Acta Hort. accepted. in press
- Laimer da Câmara Machado M., Kummert J., Candresse T., Jelkmann W.,
Cassells A., Bertaccini A., van den Heuvel J.F.J.M., Davies. D.L. 1999. Health
certification of rosaceous species based on disease-indexing of in vitro plants:
validation of diagnostics and diagnostic strategies. ISHS Conference on Methods
and Markers for Quality Assurance in Micropropagation. Cork. 24-27 August 1999.
Abstracts: 104-106.
- Lee I.M., Bertaccini A., Vibio M. and Gunderson D.E., 1995. Detection of
multiple phytoplasmas in perennial fruit trees with decline symptoms in Italy.
Phytopathol. 85: 728-735.
- Lorenz K.-H., Schneider B., Ahrens U., and Seemüller E., 1995.
Detection of apple proliferation and pear decline phytoplasmas by PCR
amplification of ribosomal and nonribosomal DNA. Phytopathol. 85: 771-776.
- MacKenzie D. J., McLean M. A., Mukerji S., and Green M., 1997. Improved
RNA extraction from woody plants for the detection of viral pathogens by
reverse transcription-polymerase chain reaction. Plant Dis. 81: 222-226.
- Malinowski T., Komorowska B., Gokis T., and Zawadzka B., 1988.
Detection of apple stem pitting virus and pear vein yellows virus using
reverse transcription - polymerase chain reaction. Acta Hort. 472.
- Marinho V.L.A., Kummert J., Rufflard G., Colinet D., and Lepoivre P., 1998.
Detection of apple stem grooving virus in dormant apple trees by using crude
extracts as templates for one-step-RT-PCR. Plant Diseases 82: 785-790.
- Nemchinov L., Hadidi A., and Faggioli F., 1998. PCR-detection of apple
stem pitting virus from pome fruit hosts and sequence variability among
viral isolates. Acta Hort. 472.
- Nemchinov L., Hadidi A., Candresse T., Foster J. A., and Verderevskaya T.,
1995. Sensitive detection of apple chlorotic leaf spot virus from infected
apple or peach tissue using RT-PCR, IC-RT-PCR, or multiplex IC-RT-PCR.
Acta Hort. 386: 51-62.
- Parakh DR., Shamloul AM., Hadidi A., Waterworth HE., Scott SW., Howell HE.
and Mink G., 1995. Detection of Prune Dwarf Virus from infected stone fruits using
reverse transcription polymerase chain reaction. Acta Horti. 386: 421 - 430.
- Powell C. A., Hadidi A., and Halbrendt J. M., 1991. Detection and
distribution of tomato ringspot virus in infected nectarine trees using
ELISA and transcribed RNA probes. HortScience 26: 1290-1292.
- Rott M., and Jelkmann W., 1999. Detection of filamentous viruses from sweet
cherry. Phytomedizin 1, 48.
- Rott M., and Jelkmann W., 1999. Detection and characterization of several
filamentous viruses from sweet cherry. XIth International Congress of Virology
9-13 Aug. 1999, 368.
- Rott M., and Jelkmann W., 1999. Charakterisierung, PCR-Diagnose und
Untersuchungen zur Verbreitung eines zweiten Closterovirus als Verursacher der
litte cherry Erkrankung an Süßkirschen. Phytomedizin 3, 64.
- Rott M. E., and Jelkmann W., 2000. Complete nucleotide sequence of cherry
necrotic rusty mottle virus. Archives of Virology (in press).
- Rott M. E., and Jelkmann W., 2000. Complete nucleotide sequence of cherry
necrotic mottle virus. Phytopathol. 90, 67.
- Rott M. E., and Jelkmann W., 2000. Detection and partial characterization of a
second viral agent associated with little cherry disease. Phythopthology 90, 67.
- Rott M. E., and Jelkmann W., 2000. Detection and partial characterization of
a second viral agent associated with little cherry disease. Acta Hort. (in press).
- Rott M. E., and Jelkmann W., 2000. Complete nucleotide sequence of cherry
necrotic rusty mottle virus. Acta Hort. (in press).
- Rott M. E., and Jelkmann W., 2000. Development of PCR primer pairs for the
characterization and detection of several related filamentous viruses of cherry.
Acta Hort. (in press).
- Rott M. E., and Jelkmann W., 2000. Molekulare Charakterisierung eines zweiten
Closterovirus assoziiert mit der Kleinfrüchtigkeit der Sübkirsche (little cherry).
Mitteilungen aus der Biologischen Bundesanstalt für Land- und Forstwirtschaft
Berlin-Dahlem (im Druck).
- Rott M. E., and Jelkmann W., 2000. Neue Erkenntnisse und Entwicklung von
Nachweisverfahren für wirtschaftlich bedeutsame und wenig beschriebene Kirschvirosen.
Mitteilungen aus der Biologischen Bundesanstalt für Land- und Forstwirtschaft
Berlin-Dahlem (im Druck).
- Rott M. E., and Jelkmann W., 2000. Charakterisierung und Nachweis filamentöser
Viren an Kirschen und Verwandtschaften zu cherry green ring mottle virus.
Phytomedizin 2, 25.
- Rowhani A., Biardi L., and Golino D. A., 1998. Detection of viruses of woody
host plants using colorimteric PCR. Acta Hort. 472.
- Rowhani A., Maningas M. A., Lile L. S., Daubert S. D., and Golino D. A.,
1995. Development of a detection system for viruses of woody plants based
on PCR analysis of immobilized virions. Phytopathol. 85: 347-352.
- Schwarz K., and Jelkmann W., 1998. Detection and characterization of
European apple stem pitting virus isolates of apple and pear by PCR and
partial sequence analysis. Acta Hort. 472.
- Smart C.D., Schneider B., Blomquist C.L., Guerra L.J., Harrison N.A.,
Ahrens U., Lorenz K.H., Seemuller E. and Kirkpatrick B.C., 1996. Phytoplasma
specific PCR primers based on sequences of the 16S - 23S rRNA spacer region.
Applied and Environmental Microbiology 62: 2988-2993.
- Spiegel S., Scott S. W., Bowman-Vance V., Tam Y., Galiakparov N. N., and
Rosner A., 1996. Improved detection of prunus necrotic ringspot virus by the
polymerase chain reaction. Eur. J. Plant Pathol. 102: 681-685.
- Veronesi F., Bertaccini A., Parente A., Mastronicola M., and Pastore M., 2000.
PCR indexing of phytoplasma-infected micropropagated periwinkle treated with
PAP-II, a ribosome inactivating protein from Phytolacca americana leaves.
Acta Hort. 530: 113-119.
- Vitushkina M., Fechtner B., AgranovskyA. A., and Jelkmann W., 1997.
Development of an RT-PCR for the detection of little cherry virus and
characterization of some isolates occuring in Europe. Eur. J. Plant Pathol.
103: 803-808.
Back to pagetop
|